Enhancing Protein Fitness with Deep Learning
Dual Degree Thesis on sequence-structure fusion using LMs and GNNs for function prediction and generative sequence design
Dual Degree Thesis | August 2023 - June 2024
Guides: Dr. Radhakrishnan Mahadevan (University of Toronto), Dr. Nirav Bhatt (IIT Madras)
Nominated for Best Thesis in Data Science Award
Overview
This thesis explores the fusion of sequence and structural information to predict protein function and generate novel sequences with enhanced properties using deep learning. The work demonstrates state-of-the-art performance on PEER and FLIP benchmarks.
Key Contributions
1. Sequence-to-Function Learning Pipeline
- Employed competitive convolutional and attention-based pooling architectures for sequence-to-function learning
- Utilized OmegaFold to predict structures for all sequences in curated datasets
- Created structure-aware graphs encoding dihedral angles, sidechains, and orientations as features
- Designed custom PyTorch data loaders for efficient processing of large-scale protein datasets
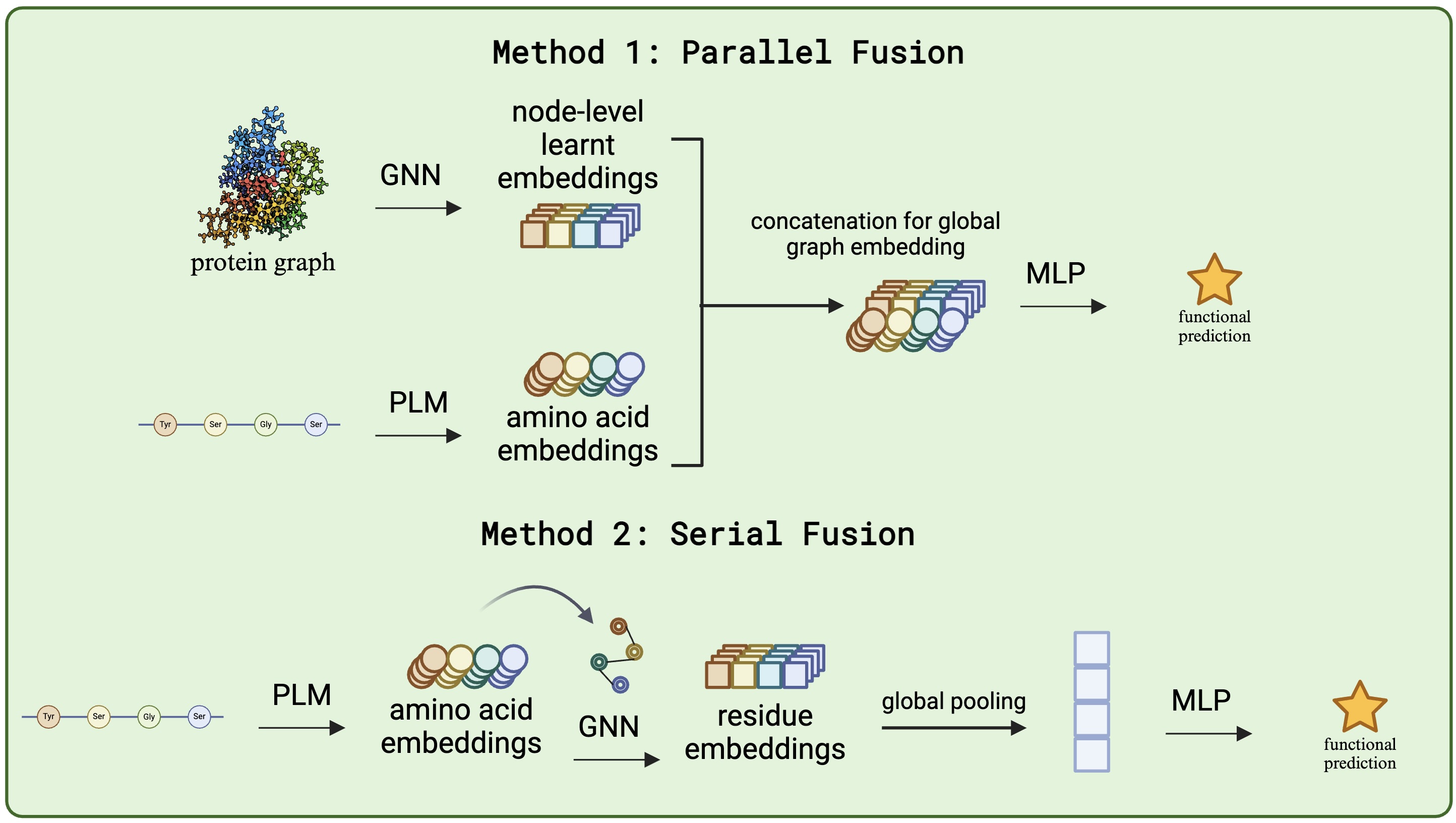
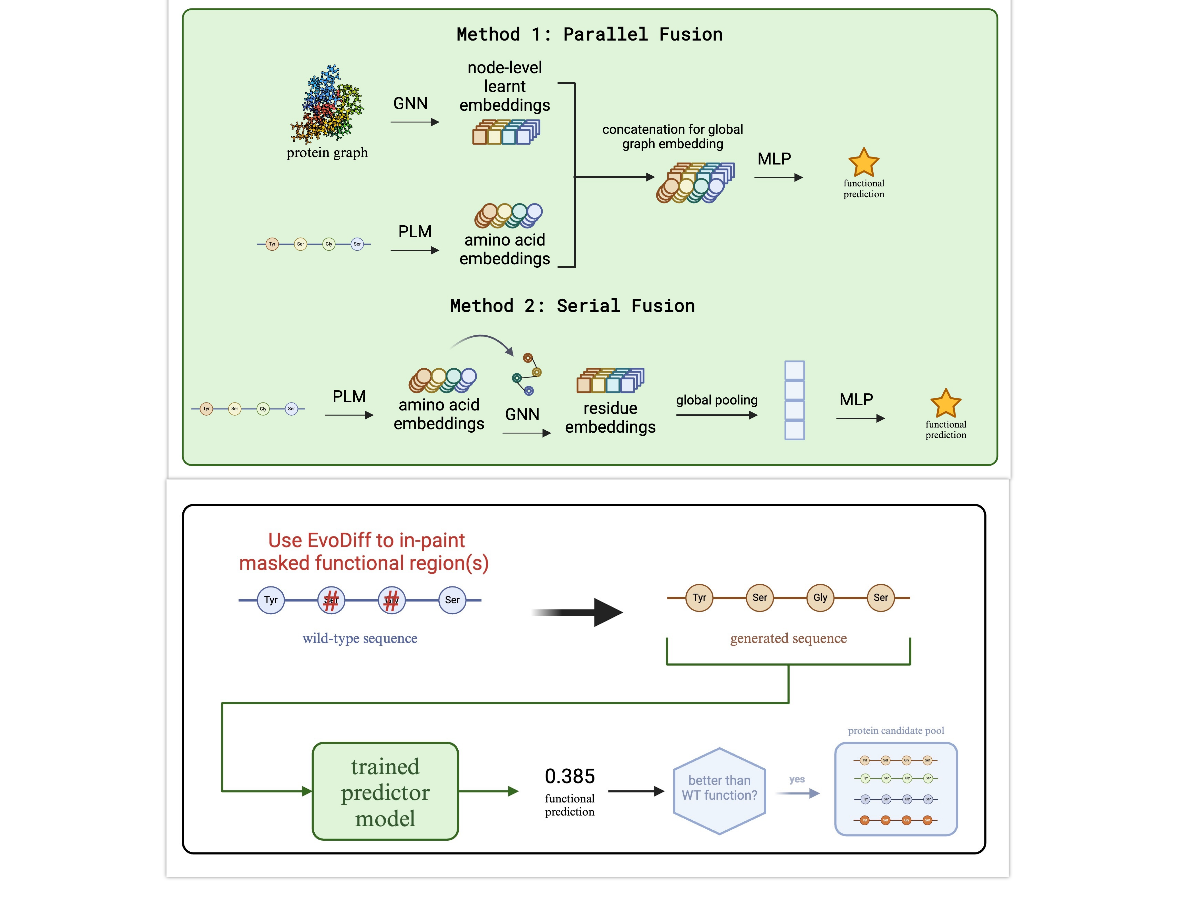
2. Graph Neural Network Architecture
- Demonstrated superior GNN performance over sequence-only approaches through fusion of ESM-2 embeddings with structural information
- Implemented parallel and cross-attention-based fusion mechanisms for benchmarking
- Introduced a vector gating mechanism creating dependencies between scalar and vector features
- Achieved state-of-the-art performance on PEER and FLIP benchmarks
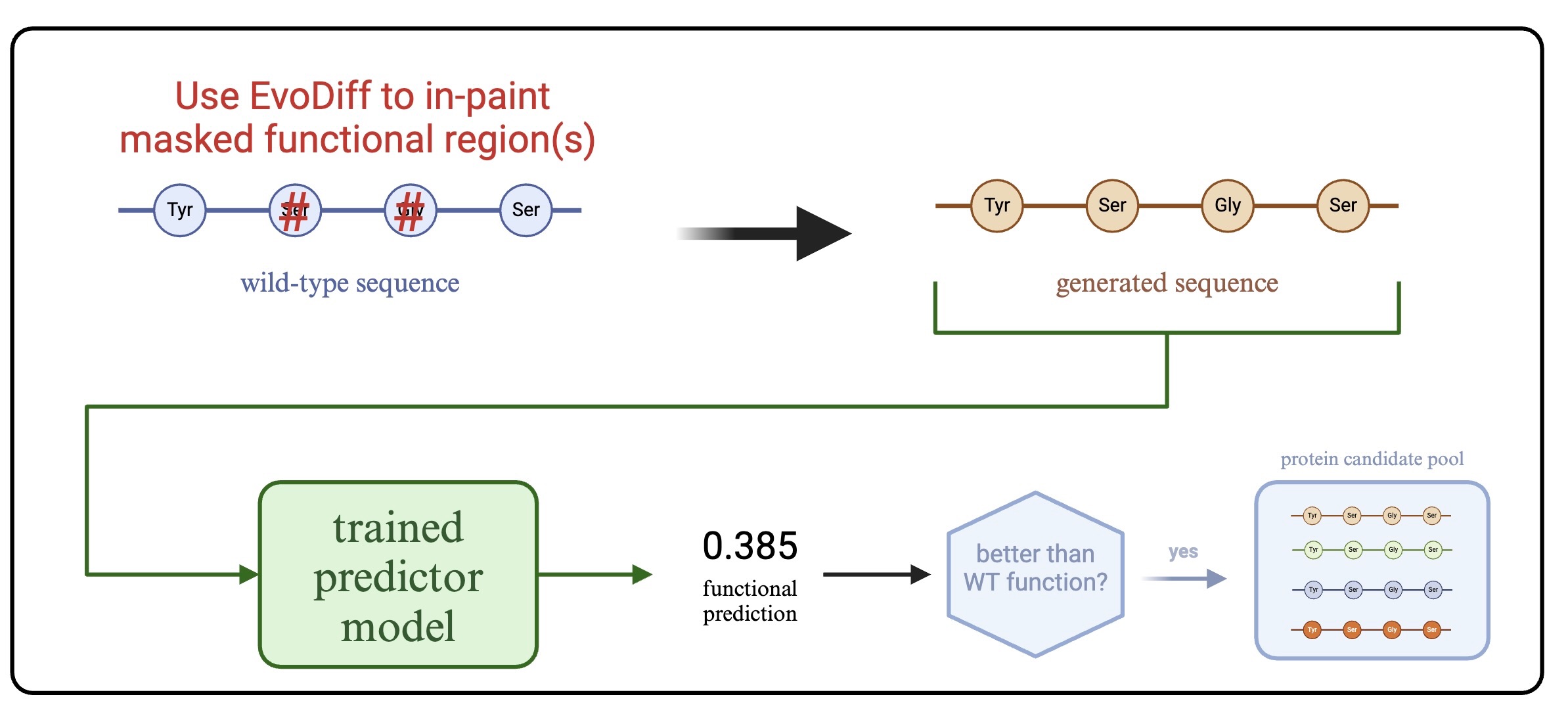
3. Generative Design Strategy
Our approach for designing enhanced protein variants:
- Masking: Mask functional residues or active site domains of wildtype proteins
- Generation: Use EvoDiff’s Order-Agnostic Diffusion Model (OADM) to generate novel sequences via inpainting
- Filtering: Generate structures with OmegaFold and discard sequences with pLDDT < 70
- Screening: Apply trained GNN to predict functional values and identify candidates with enhanced function
Experimental Results
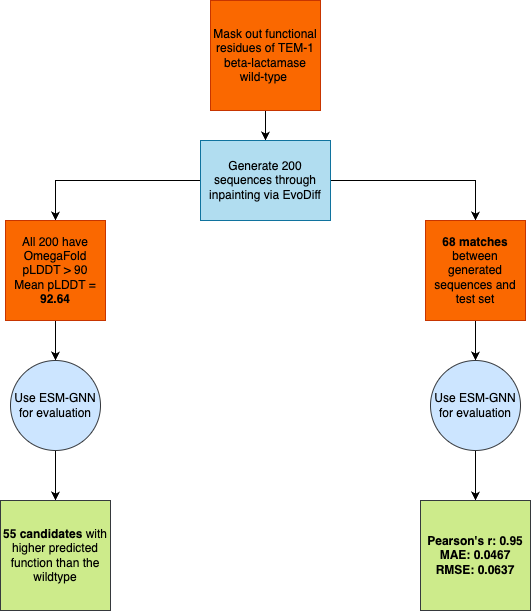
β-lactamase (TEM-1)
- Generated 200 novel sequences by inpainting masked functional regions (residues 103-105, 168-170, 238-241)
- All 200 sequences achieved pLDDT > 90 (average: 92.64), demonstrating EvoDiff’s capability for structurally plausible generation
- 55 sequences predicted to exhibit higher function than wildtype
- Identified 68 matches with PEER benchmark test set, validating ESM-GNN’s predictive efficacy
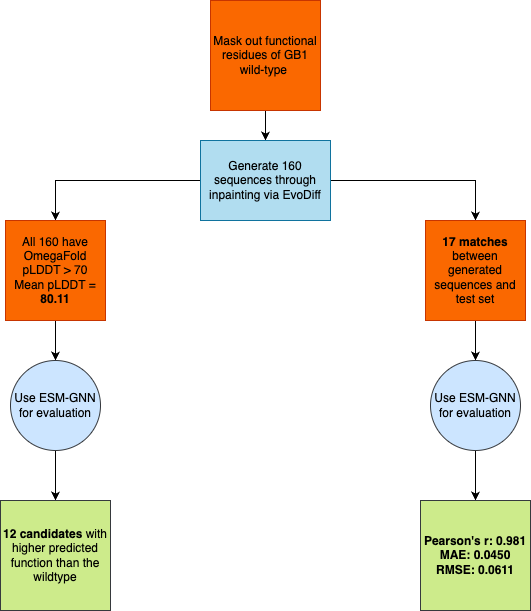
GB1 Protein
- Generated 160 novel sequences by masking 4 functional sites (V39, D40, G41, V54)
- All sequences achieved pLDDT > 70 (average: 80.11)
- 12 sequences predicted to have higher function than wildtype
- Identified 17 matches with test set; successfully identified all 10 true enhanced-function candidates
- Demonstrated high reliability for preliminary screening before experimental validation
Technical Details
Models: ESM-2, OmegaFold, EvoDiff, Custom GNN with vector gating, GVP-GNN
Frameworks: PyTorch, PyTorch Geometric
Benchmarks: PEER, FLIP
Key Innovation: Fusion of sequence embeddings with structure-aware graph representations
Impact
This work demonstrates that combining protein language models with graph neural networks can:
- Accurately predict function from sequence and structure
- Generate novel sequences with enhanced properties
- Significantly reduce experimental validation costs through computational screening
This research was presented at Machine Learning in Computational Biology (MLCB) 2024:
References
2024
- MLCB
 Screening Protein Sequences Generated via Conditional Diffusion for Enhanced Fitness using a GNN-based Function PredictorIn Machine Learning for Computational Biology (MLCB), 2024Poster presentation
Screening Protein Sequences Generated via Conditional Diffusion for Enhanced Fitness using a GNN-based Function PredictorIn Machine Learning for Computational Biology (MLCB), 2024Poster presentation